4. Ensembles
4.1. Ensemble reduction
This tutorial will explore ensemble reduction (also known as ensemble selection) using xscen. This will use pre-computed annual mean temperatures from xclim.testing.
[2]:
from xclim.testing import open_dataset
import xscen as xs
datasets = {
"ACCESS": "EnsembleStats/BCCAQv2+ANUSPLIN300_ACCESS1-0_historical+rcp45_r1i1p1_1950-2100_tg_mean_YS.nc",
"BNU-ESM": "EnsembleStats/BCCAQv2+ANUSPLIN300_BNU-ESM_historical+rcp45_r1i1p1_1950-2100_tg_mean_YS.nc",
"CCSM4-r1": "EnsembleStats/BCCAQv2+ANUSPLIN300_CCSM4_historical+rcp45_r1i1p1_1950-2100_tg_mean_YS.nc",
"CCSM4-r2": "EnsembleStats/BCCAQv2+ANUSPLIN300_CCSM4_historical+rcp45_r2i1p1_1950-2100_tg_mean_YS.nc",
"CNRM-CM5": "EnsembleStats/BCCAQv2+ANUSPLIN300_CNRM-CM5_historical+rcp45_r1i1p1_1970-2050_tg_mean_YS.nc",
}
for d in datasets:
ds = open_dataset(datasets[d]).isel(lon=slice(0, 4), lat=slice(0, 4))
ds = xs.climatological_op(
ds,
op="mean",
window=30,
periods=[[1981, 2010], [2021, 2050]],
horizons_as_dim=True,
).drop_vars("time")
datasets[d] = xs.compute_deltas(ds, reference_horizon="1981-2010")
ERROR 1: PROJ: proj_create_from_database: Open of /home/docs/checkouts/readthedocs.org/user_builds/xscen/conda/latest/share/proj failed
4.1.1. Preparing the data
Ensemble reduction is built upon climate indicators that are relevant to represent the ensemble’s variability for a given application. In this case, we’ll use the mean temperature delta between 2021-2050 and 1981-2010, but monthly or seasonal indicators could also be required. The horizons_as_dim argument in climatological_op can help combine indicators of multiple frequencies into a single dataset. Alternatively, xscen.utils.unstack_dates can also accomplish the same thing if the
climatological operations have already been computed.
The functions implemented in xclim.ensembles._reduce require a very specific 2-D DataArray of dimensions “realization” and “criteria”. The first solution is to first create an ensemble using xclim.ensembles.create_ensemble, then pass the result to xclim.ensembles.make_criteria. Alternatively, the datasets can be passed directly to xscen.ensembles.reduce_ensemble and the necessary preliminary steps will be accomplished automatically.
In this example, the number of criteria will corresponds to: indicators x horizons x longitude x latitude, but criteria that are purely NaN across all realizations will be removed.
Note that xs.spatial_mean could have been used prior to calling that function to remove the spatial dimensions.
4.1.2. Selecting a reduced ensemble
NOTE
Ensemble reduction in xscen is built upon xclim.ensembles. For more information on basic usage and available methods, please consult their documentation.
Ensemble reduction through xscen.reduce_ensemble consists in a simple call to xclim. The arguments are: - data, which is the aforementioned 2D DataArray, or the list/dict of datasets required to build it. - method is either kkz or kmeans. See the link above for further details on each technique. - horizons is used to instruct on which horizon(s) to build the data from, if data needs to be constructed. - create_kwargs, the arguments to pass to
xclim.ensembles.create_ensemble if data needs to be constructed. - kwargs is a dictionary of arguments to send to the clustering method chosen.
[3]:
selected, clusters, fig_data = xs.reduce_ensemble(
data=datasets, method="kmeans", horizons=["2021-2050"], max_clusters=3
)
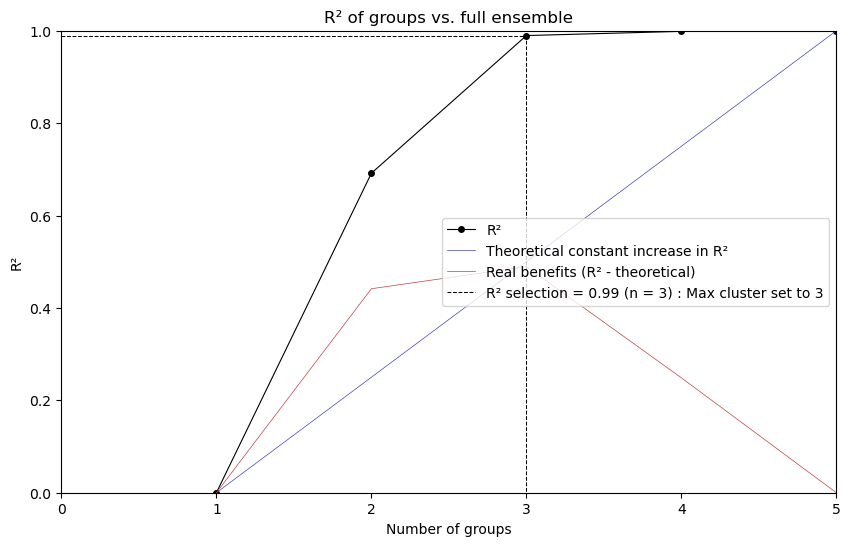
The method always returns 3 outputs (selected, clusters, fig_data): - selected is a DataArray of dimension ‘realization’ listing the selected simulations. - clusters (kmeans only) groups every realization in their respective clusters in a python dictionary. - fig_data (kmeans only) can be used to call xclim.ensembles.plot_rsqprofile(fig_data)
[4]:
selected
[4]:
<xarray.DataArray 'realization' (realization: 3)> Size: 96B
array(['ACCESS', 'BNU-ESM', 'CNRM-CM5'], dtype='<U8')
Coordinates:
* realization (realization) <U8 96B 'ACCESS' 'BNU-ESM' 'CNRM-CM5'
Attributes:
axis: E[5]:
# To see the clusters in more details
clusters
[5]:
{0: <xarray.DataArray 'realization' (realization: 3)> Size: 96B
array(['ACCESS', 'CCSM4-r1', 'CCSM4-r2'], dtype='<U8')
Coordinates:
* realization (realization) <U8 96B 'ACCESS' 'CCSM4-r1' 'CCSM4-r2'
Attributes:
axis: E,
1: <xarray.DataArray 'realization' (realization: 1)> Size: 32B
array(['BNU-ESM'], dtype='<U8')
Coordinates:
* realization (realization) <U8 32B 'BNU-ESM'
Attributes:
axis: E,
2: <xarray.DataArray 'realization' (realization: 1)> Size: 32B
array(['CNRM-CM5'], dtype='<U8')
Coordinates:
* realization (realization) <U8 32B 'CNRM-CM5'
Attributes:
axis: E}
[6]:
from xclim.ensembles import plot_rsqprofile
plot_rsqprofile(fig_data)
4.2. Ensemble partition
This tutorial will show how to use xscen to create the input for xclim partition functions.
[7]:
# Get catalog
from pathlib import Path
output_folder = Path().absolute() / "_data"
cat = xs.DataCatalog(str(output_folder / "tutorial-catalog.json"))
# create a dictionnary of datasets wanted for the partition
input_dict = cat.search(variable="tas", member="r1i1p1f1").to_dataset_dict(
xarray_open_kwargs={"engine": "h5netcdf"}
)
--> The keys in the returned dictionary of datasets are constructed as follows:
'id.domain.processing_level.xrfreq'
From a dictionary of datasets, the function creates a dataset with new dimensions in partition_dim(["source", "experiment", "bias_adjust_project"], if they exist). In this toy example, we only have different experiments. - By default, it translates the xscen vocabulary (eg. experiment) to the xclim partition vocabulary (eg. scenario). It is possible to pass rename_dict to rename the dimensions with other names. - If the inputs are not on the same grid, they can be regridded
through regrid_kw or subset to a point through subset_kw. The functions assumes that if there are different bias_adjust_project, they will be on different grids (with all source on the same grid). If there is one or less bias_adjust_project, the assumption is thatsource have different grids. - You can also compute indicators on the data if the input is daily. This can be especially useful when the daily input data is on different calendars.
[8]:
# build a single dataset
import xclim as xc
ds = xs.ensembles.build_partition_data(
input_dict,
subset_kw=dict(name="mtl", method="gridpoint", lat=[45.5], lon=[-73.6]),
indicators_kw={"indicators": [xc.atmos.tg_mean]},
)
ds
[8]:
<xarray.Dataset> Size: 220B
Dimensions: (time: 2, scenario: 4, model: 1)
Coordinates:
* time (time) datetime64[ns] 16B 2001-01-01 2002-01-01
* scenario (scenario) object 32B 'ssp126' 'ssp245' 'ssp370' 'ssp585'
* model (model) <U23 92B 'NCC_NorESM2-MM_r1i1p1f1'
lat float64 8B 45.0
lon int64 8B -74
Data variables:
tg_mean (scenario, model, time) float64 64B dask.array<chunksize=(4, 1, 2), meta=np.ndarray>
Attributes: (12/22)
comment: This is a test file created for the xscen tutori...
version: v20191108
intake_esm_vars: ['tas']
cat:id: CMIP6_ScenarioMIP_NCC_NorESM2-MM_ssp370_r1i1p1f1...
cat:type: simulation
cat:processing_level: indicators
... ...
cat:date_start: 2001-01-01 00:00:00
cat:date_end: 2002-12-31 00:00:00
cat:_data_format_: nc
cat:path: /home/docs/checkouts/readthedocs.org/user_builds...
intake_esm_dataset_key: CMIP6_ScenarioMIP_NCC_NorESM2-MM_ssp370_r1i1p1f1...
history: [2024-04-26 15:27:36] gridpoint spatial subsetti...Pass the input to an xclim partition function.
[10]:
# compute uncertainty partitionning
mean, uncertainties = xc.ensembles.hawkins_sutton(ds.tg_mean)
uncertainties
[10]:
<xarray.DataArray (uncertainty: 4, time: 4)> Size: 128B
dask.array<concatenate, shape=(4, 4), dtype=float64, chunksize=(1, 2), chunktype=numpy.ndarray>
Coordinates:
lat float64 8B 45.0
lon int64 8B -74
* time (time) object 32B 2001-01-01 00:00:00 ... 2004-01-01 00:00:00
* uncertainty (uncertainty) object 32B 'variability' 'model' ... 'total'
Attributes:
model: ['NCC_NorESM2-MM_r1i1p1f1']
scenario: ['ssp126' 'ssp245' 'ssp370' 'ssp585']NOTE
Note that the figanos library provides a function fg.partition to plot the uncertainties.